- About Us
-
CRO Services
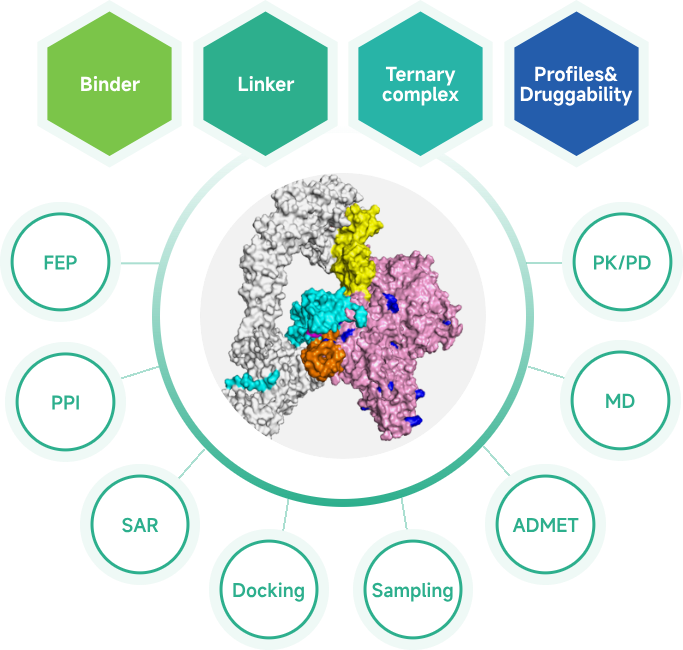
- PROTAC/Molecular Glue Services
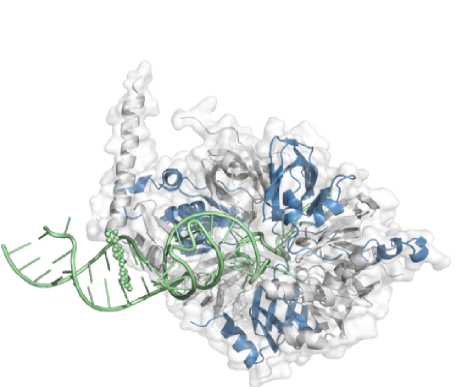
- Protein Preparation and Ternary Complex Structure Determination
- PROTAC/Molecular Glues Screening
- PROTAC Ternary Complex Kinetics (SPR)
- PROTAC Degradation Assays and Ternary Complex Assays
- PROTAC Molecule Design and Synthesis
- ADME & PK/PD Studies of PROTAC Molecules
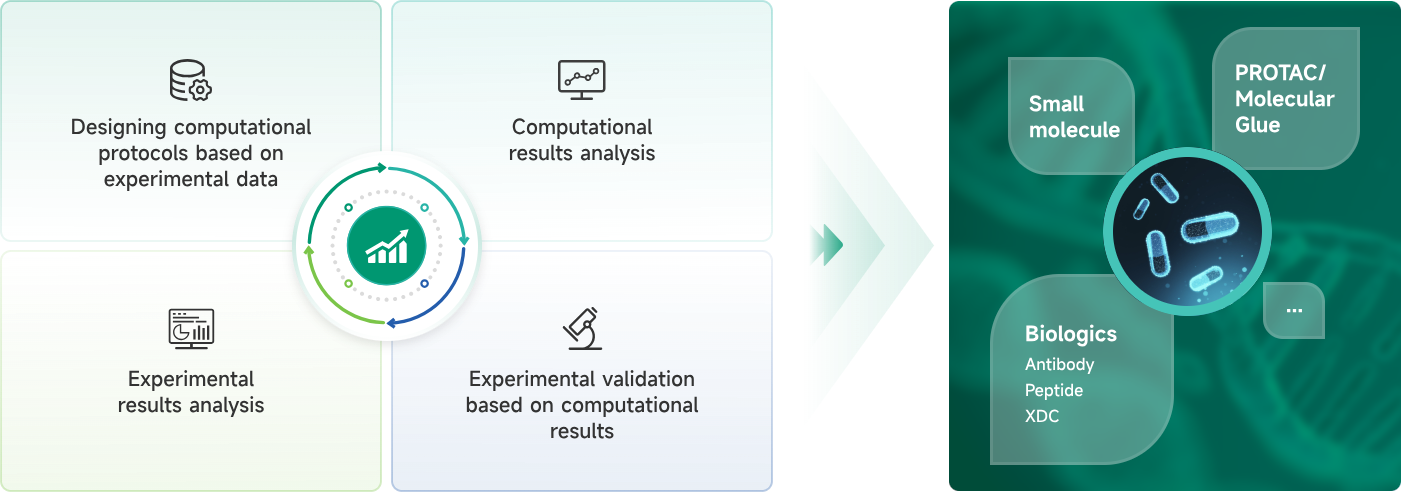
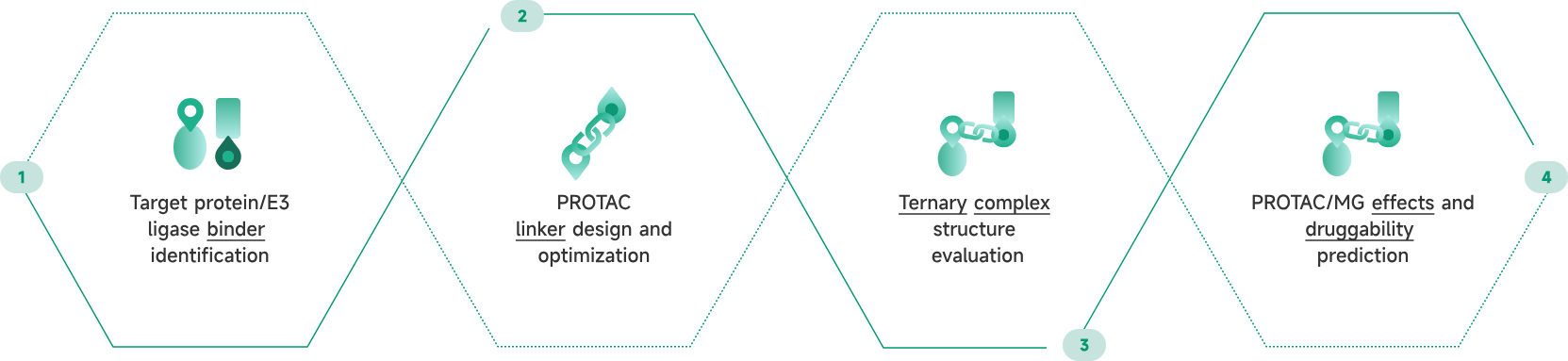
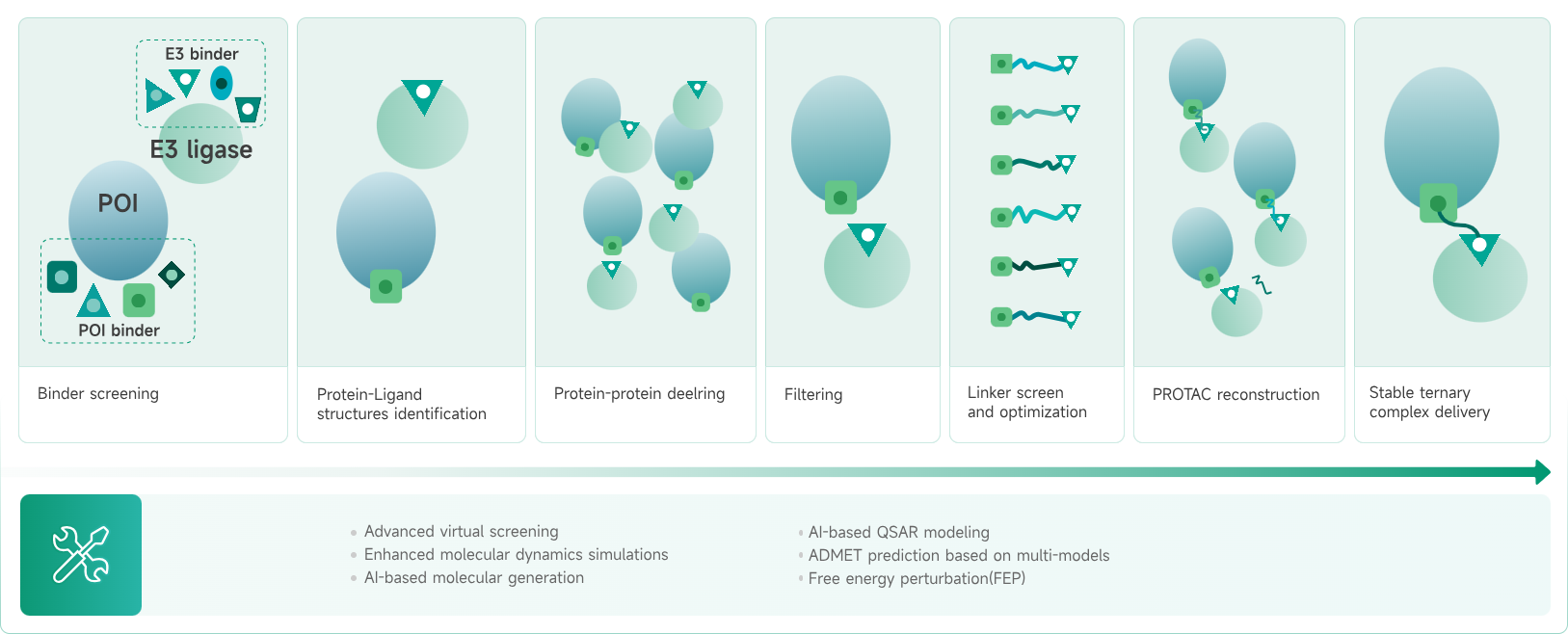
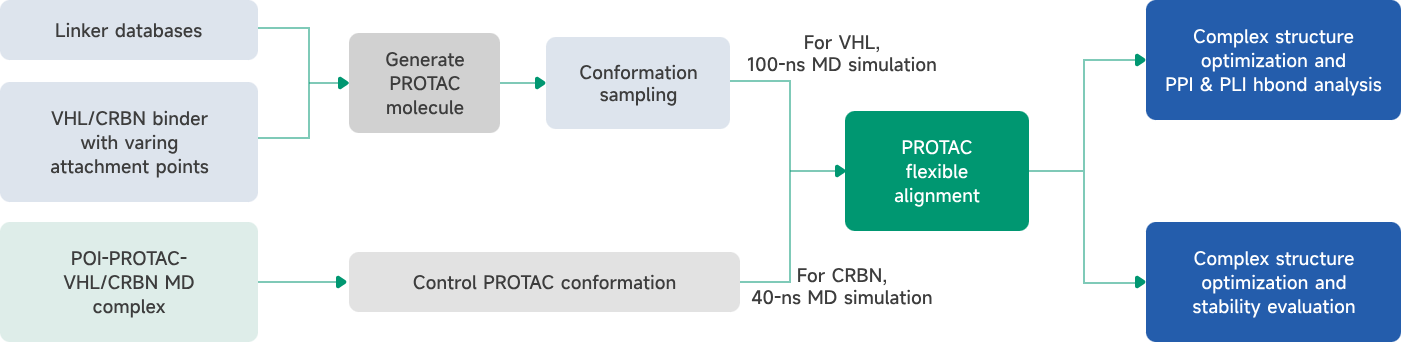
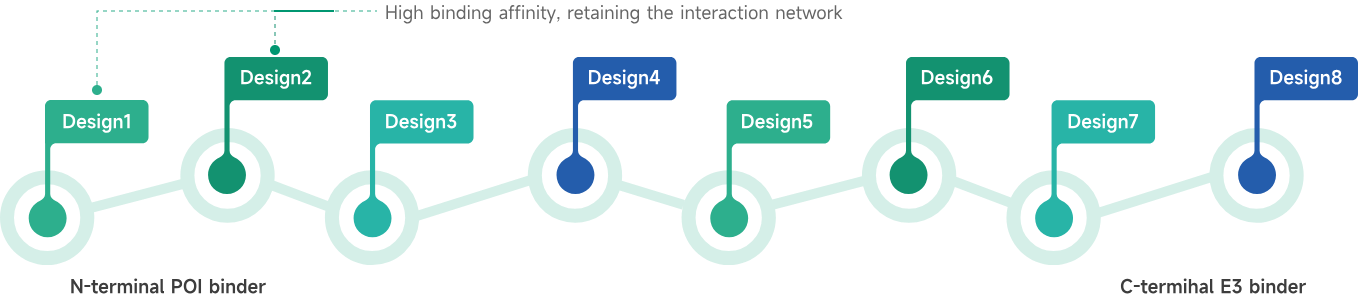
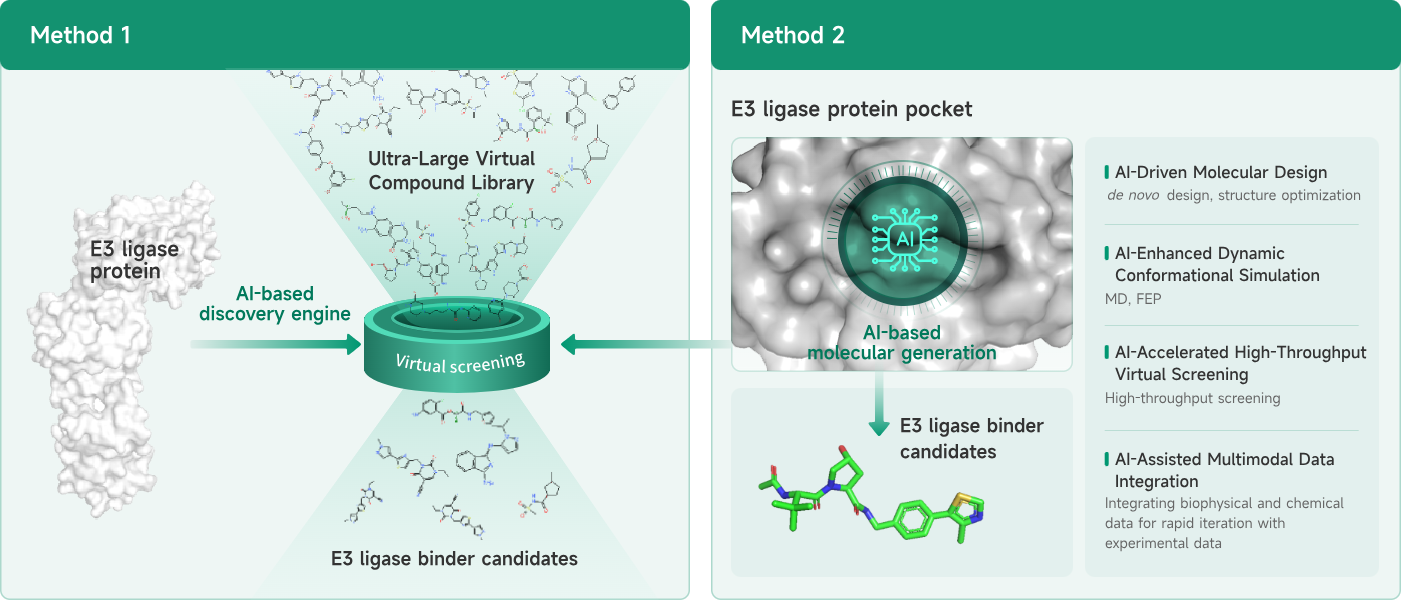
- AIDD/CADD PROTAC Design
- CDMO Services
- EFS Business
- News
- Careers
- Investor Relations
- Contact Us