
- About Us
-
CRO Services
- PROTAC/Molecular Glue Services
- Protein Preparation and Ternary Complex Structure Determination
- PROTAC/Molecular Glues Screening
- PROTAC Ternary Complex Kinetics (SPR)
- PROTAC Degradation Assays and Ternary Complex Assays
- PROTAC Molecule Design and Synthesis
- ADME & PK/PD Studies of PROTAC Molecules
- AIDD/CADD PROTAC Design
- CDMO Services
- EFS Business
- News
- Careers
- Investor Relations
- Contact Us
Fragments screening using crystal soaking is an important method in fragment-based drug discovery. This technology usually requires the resolution of the crystal structure of the drug target protein to be higher than 2.5A. In the experimental process, a single crystal of target protein usually grows first and is soaked in the solution drop with compound for a period, and then the protein crystal will be taken out with cryoprotectant and frozen in liquid nitrogen. Then the protein crystals will be shipped to the synchrotron radiation light sources for data collection. Finally, Structures will be determined by Viva scientists, and reports, structure files, as well as theraw and processed data, will be delivered.
High throughput soaking experiments:

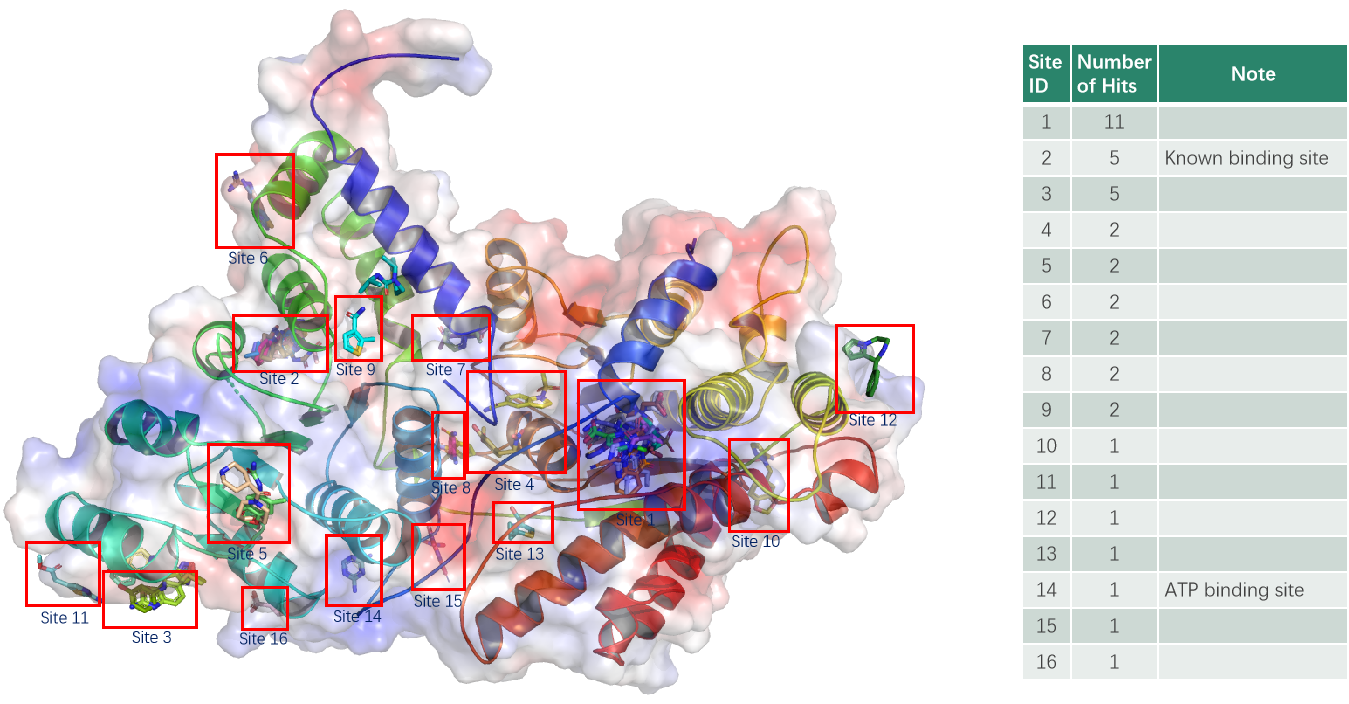
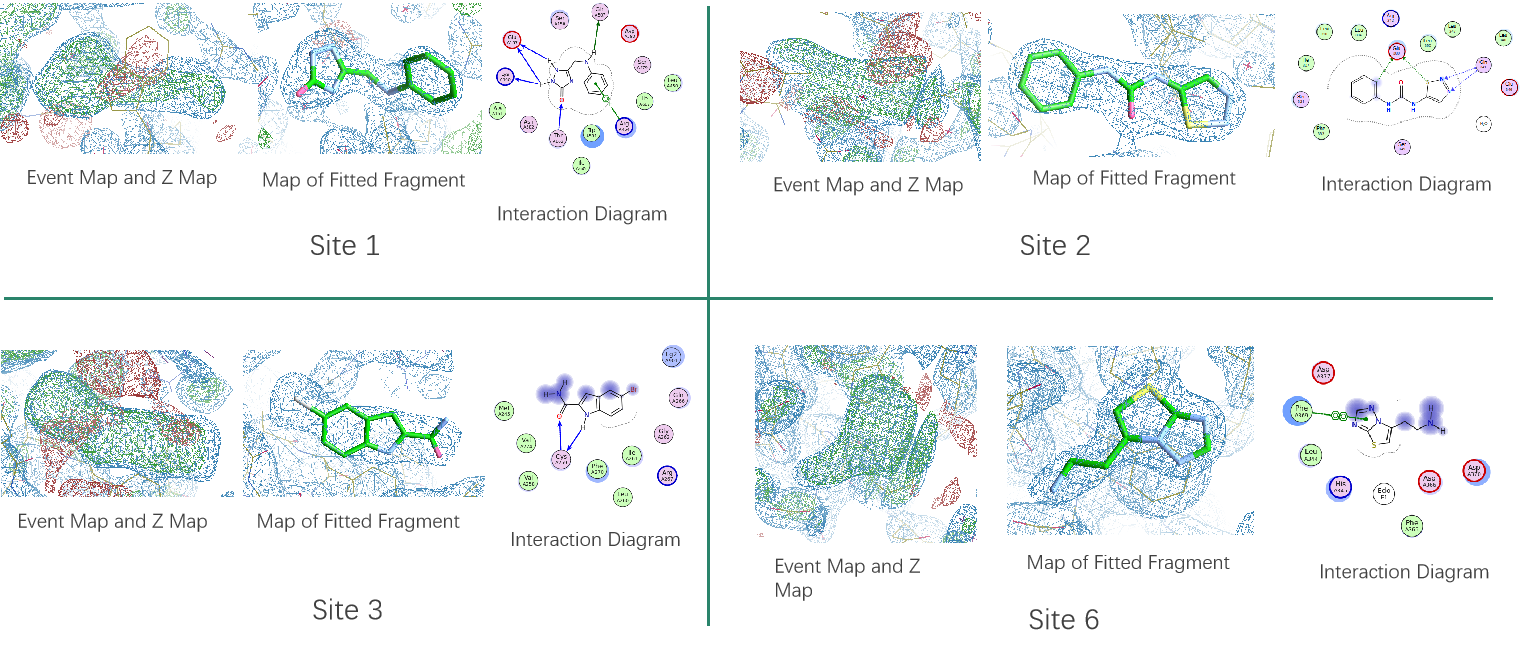
Case Study: Crystallographic fragment screening on a DNA helicase
A DNA helicase construct, comprised of ATP binding and C-terminal domain, with an ATP binding site and a known positive compound binding site, has an amino acid sequence length of 633 and a molecular weight of 76602 Da.
The Viva R&D team use vapor diffusion method to grow protein Apo crystals (with typical diffraction resolution of 2.2Å) in PEG based condition, and then collected 258 datasets after immersing the 280 crystals grown with viva's own fragment compound library of 960 compounds for 16 hours. Datasets with resolution worse than 3 Å were discarded, then 244 datasets were put into PanDDA analysis. And in this experiment, a total of 11 binding sites were screened and the fragment compound library of 960 compounds had a hit rate of 4.3%.
(The First One)
(The Second One)
Fragmented compound library of 960 compounds
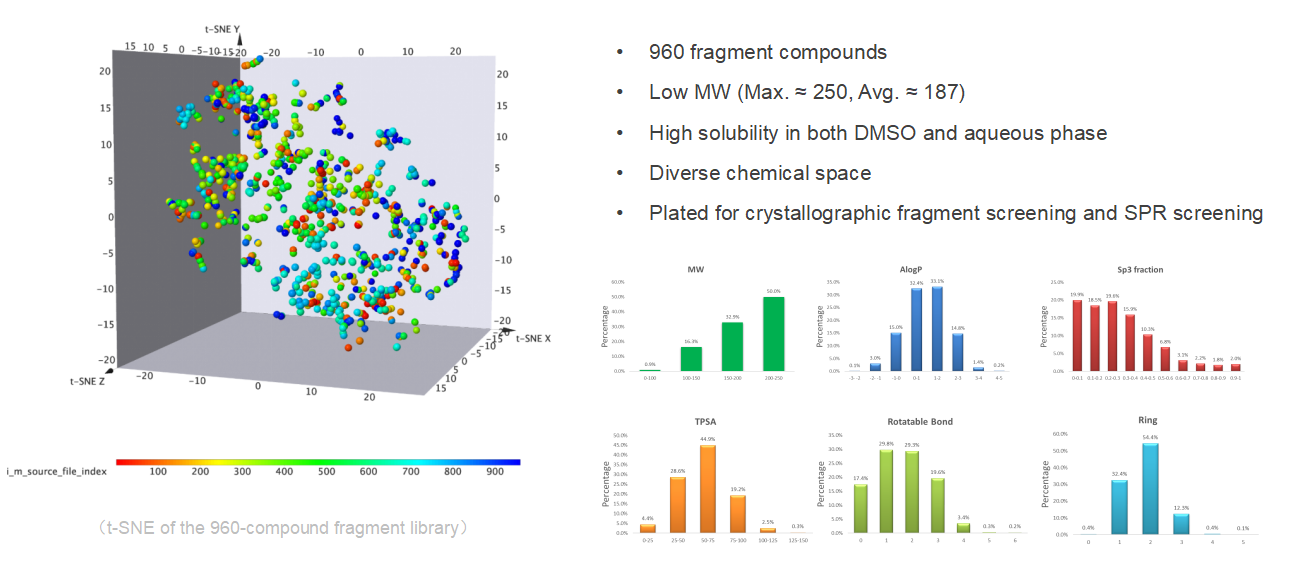
(The First One)
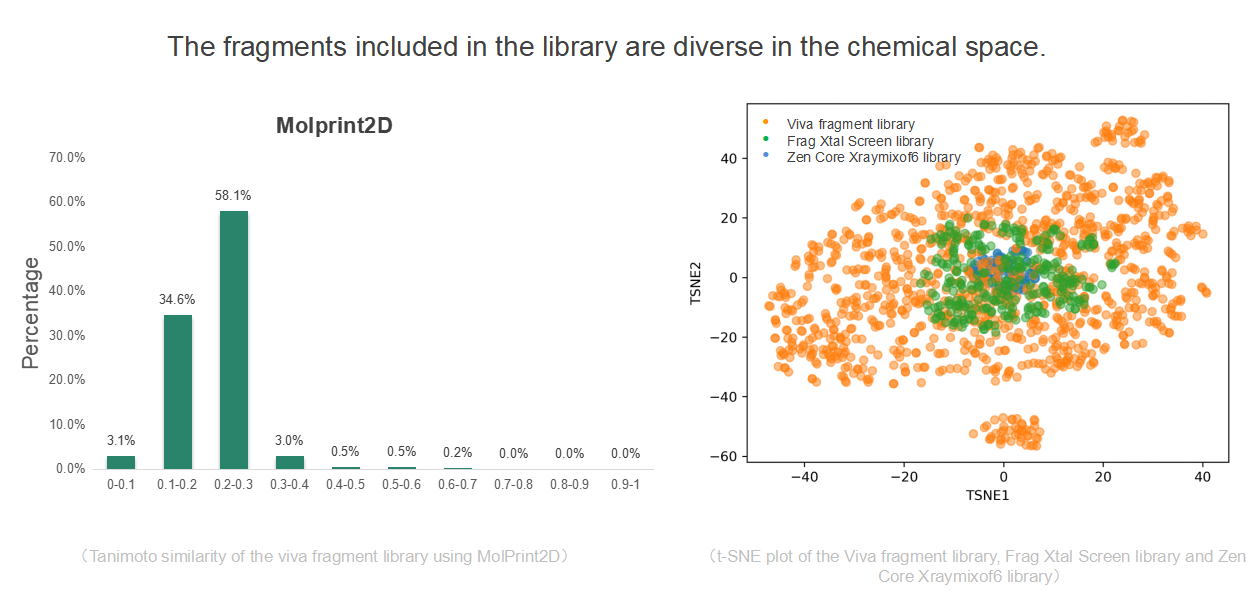
(The Second One)
Structure Gallery

Copyright © Viva Biotech All Rights Reserved. 沪ICP备19036061号
- About Us
-
CRO Services
BackCRO ServicesService & Technology
- CDMO Services
- EFS Business
- News
- Careers
- Investor Relations
- Contact Us