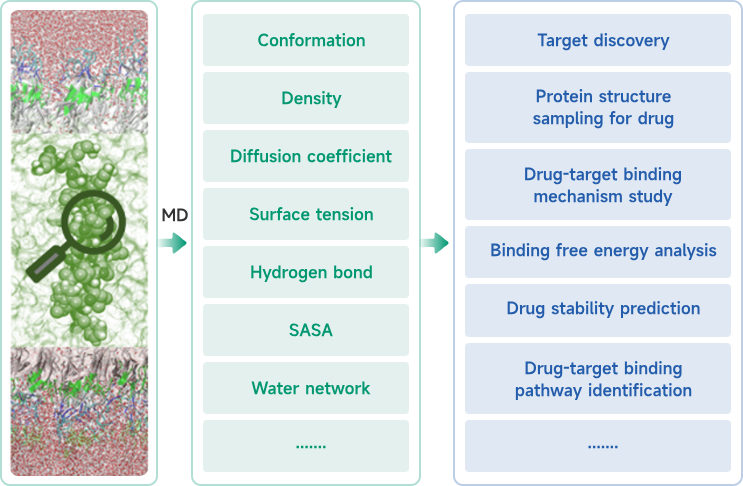
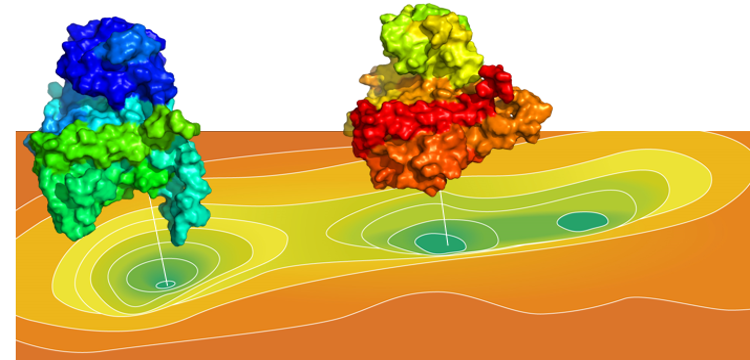
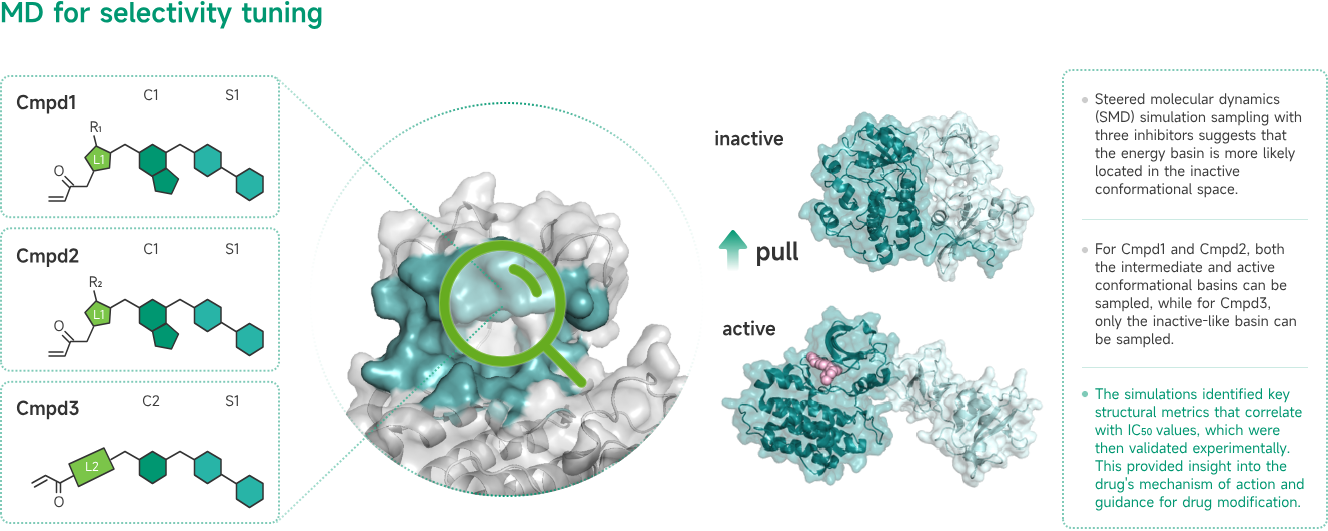
Nature Oncogene Publishes Study Highlighting TMBIM6's Transformative Potential in Treating Cancer and Beyond—Viva Biotech's AIDD/CADD Platform Made Key Contributions to the Novel Scientific Discovery
On November 29, MicroQuin published an advanced study in the Nature Portfolio titled “TMBIM6/BI-1 is an intracellular environmental regulator that induces paraptosis in cancer via ROS and Calcium-activated ERAD II pathways”. The article unveils the how the transmembrane B cell lymphoma 2-associated X protein (BAX) inhibitor motif-containing TMBIM6 can treat a variety of cancers. TMBIM6 as an IntraCellular Environmental (ICE) regulator induces paraptosis via noncanonical routes — ERAD II mechanisms. TMBIM6 agonism can induces rapid cancer cell death with no toxicity, representing an attractive therapeutic target across various diseases and injuries, including cancer. Dr. Yue Qian, Executive Director of the Computational Chemistry and Artificial Intelligence for Drug Discovery (AIDD/CADD) Platform at Viva Biotech, Hai Liu, Project Manager of the AIDD/CADD platform, and Dr. David Taddei, Vice President of Medicinal Chemistry provided computational insights to the function-related protein dynamics under different conditions. The in silico modeling made significant contributions to understanding the mechanism of action and experimental design.

(Source: Nature Oncogene website)
TMBIM 6 has been heavily researched for its cytoprotective functions. It's functional diversity includes modulating cell survival, stress, metabolism, etc. Clinical research shows TMBIM6 plays a key role in many of the world's top diseases/injuries i.e., Alzheimer's, Parkinson's, and cancer, where TMBIM6 expression impacts patient survival, chemoresistance, cancer progression, and metastasis. The study shows TMBIM6 is activated by, and undergoes, different conformational changes that dictate its function following a significant change in the cell’s ICE. In cancer cells TMBIM6 agonism results in rapid paraptotic induction irrespective of the cancer type, sub-type, genotype or phenotype. Furthermore, the level of TMBIM6 expression in cancer did not dictate the level of paraptotic induction; however, it did dictate the rate at which paraptosis occurred. Instead, TMBIM6 agonism in cancer upregulates cytosolic Ca2+ and reactive oxygen species (ROS), activates lysosome biogenesis, and induces paraptosis via ERAD II mechanisms. In xenograft models, TMBIM6 agonism induces rapid cancer cell death with no toxicity, even at high doses of TMBIM6 agonist (>450 mg/kg). In summary, this study shows TMBIM6's functional diversity is only activated by severe ICE change in diseased/injured cells, highlighting its transformative potential as a therapeutic target across various diseases and injuries, including cancer.
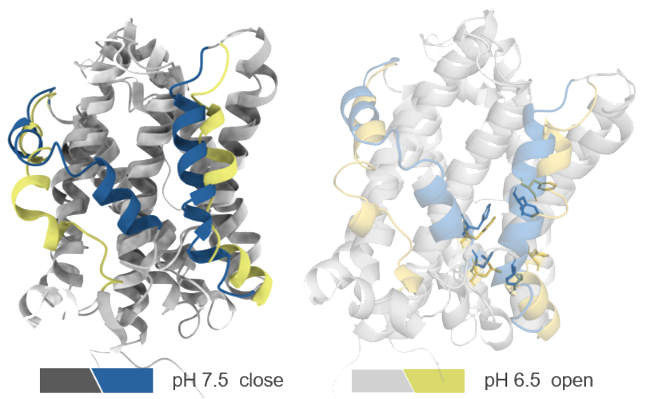
TMBIM6's functional diversity is key in the pathogenesis of many diseases including neurodegenerative disease, ischemic-reperfusion injury, and cancer, etc. The complexity and diversity of TMBIM6's functions pose considerable challenges for researchers to understand its structure and function. TMBIM6 is capable of detecting changes in the cell's intracellular environment (ICE) and reacts with a conformational change, and environmental stimuli (pH, Ca2+, and ROS) also impact on TMBIM6 conformation. The deep understanding of TMBIM6 dynamic behavior is crucial for developing TMBIM6-based therapeutic strategy.
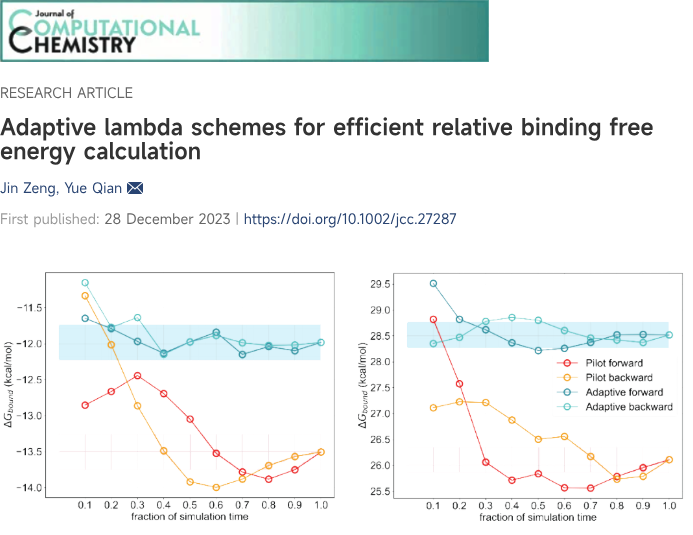
In order to investigate the TMBIM6's capabilities in detecting unique ICE change that induce conformational change to activate specific TMBIM6 functions, Viva Biotech's AIDD/CADD team utilized structural modeling, optimization and constant pH molecular dynamic simulation (CpHMD) to explore TMBIM6's conformational change. Despite that the TMBIM6 structure has not been experimentally determined, the team is able to identify significant changes in the transmembrane region and alpha-helical formation of TMBIM6 under different pH conditions. The CpHMD results fully align with the circular dichroism data (CD) in terms of the secondary structure propensity. This data further supports the idea that TMBIM6 can detect ICE change, which induces structural conformational change. This approach provided insights into how molecular structural changes influence biological functions. By accurately simulating the protein's behavior in dynamic environments and analyzing the roles of key residues, the team established a solid theoretical and structural foundation for TMBIM6-targeted drug design. These findings demonstrate our AIDD/CADD team's advanced capabilities in exploring novel targets and mechanisms in drug discovery.

(Source: Oncogene)
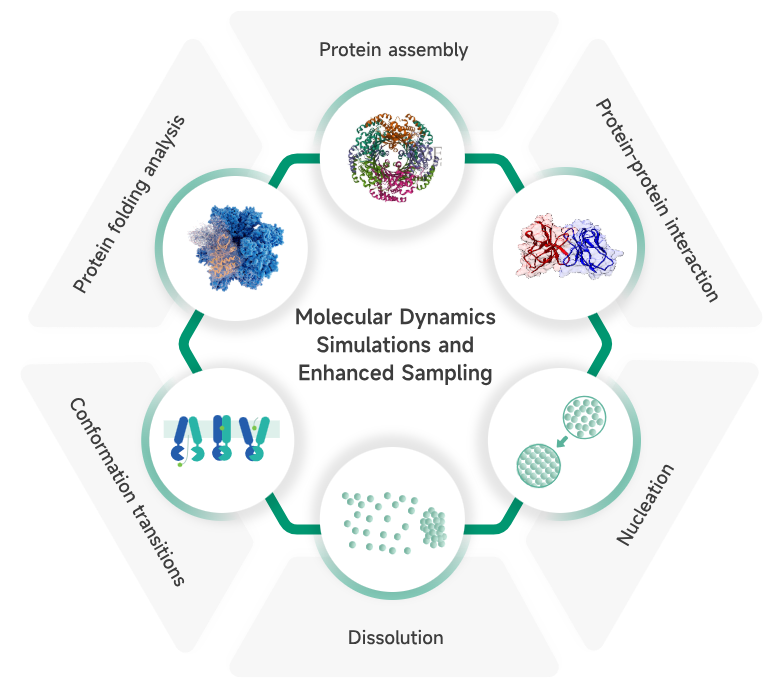
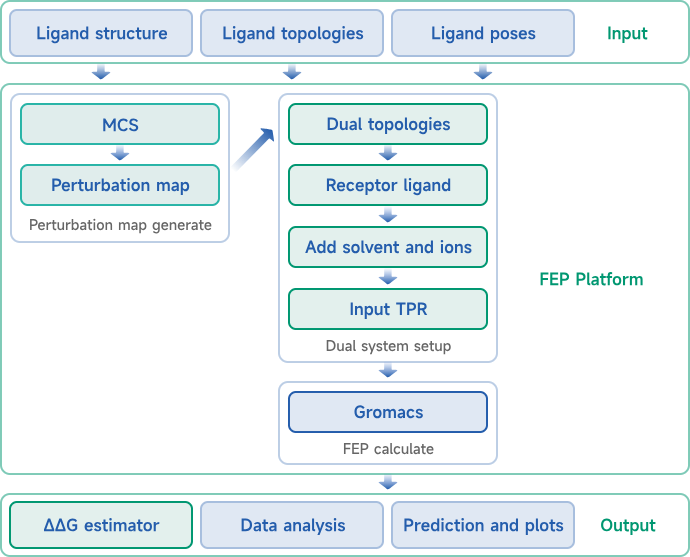
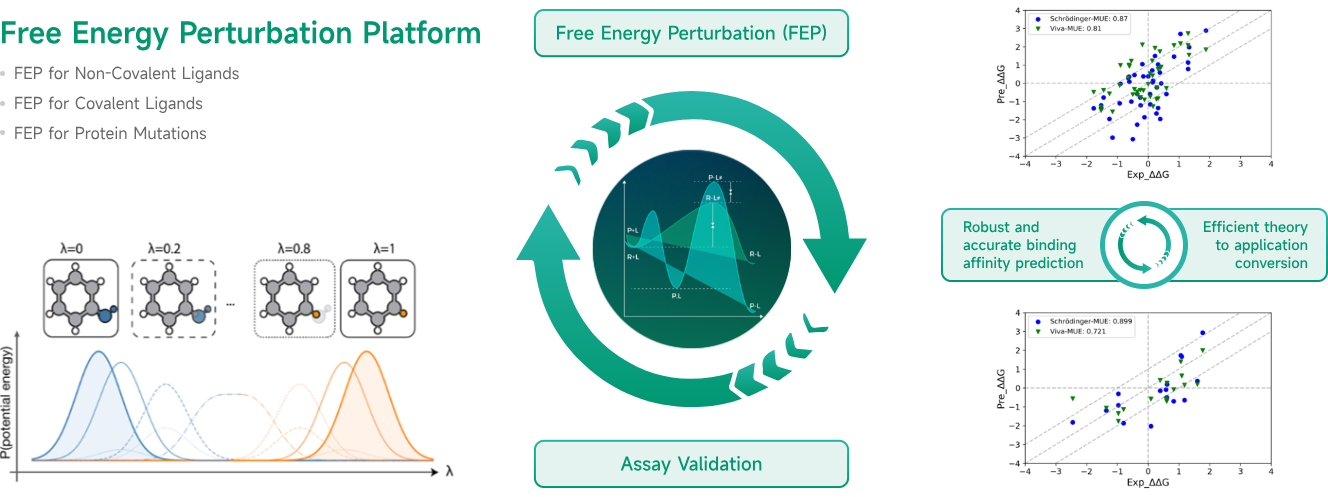
At Viva Biotech, Dr. Yue Qian has established a team of talents that represents the fast-evolving scientific fronts and diverse demographic backgrounds. This team represents a comprehensive AI-driven drug discovery platform that integrates cutting-edge computational methods with real-world experimental observations. Techniques include virtual screening, de novo design, molecular dynamics, free energy perturbation, and a series of deep-learning based algorithms, covering the drug discovery process from target identification to lead optimization and to preclinical research. The team puts together extensive experience in developing a range of drug modalities such as small molecules, antibodies, peptides, and RNA-targeting small molecules. We collaborate seamlessly between the modeling team and the experimental validations, which significantly reduces both the time and cost associated with drug discovery while offering forward-thinking solutions to address critical challenges in pharmaceutical research. With the rapid technological advancement, Viva Biotech harnesses the synergy of AI and experimental technologies to unlock new possibilities in drug discovery and drive the biopharmaceutical industry toward greater precision and efficiency.
For further details on the paper, please refer to:
Robinson, K.S., Sennhenn, P., Yuan, D.S. et al. TMBIM6/BI-1 is an intracellular environmental regulator that induces paraptosis in cancer via ROS and Calcium-activated ERAD II pathways. Oncogene (2024). https://doi.org/10.1038/s41388-024-03222-x
For more information about Viva Biotech's AIDD/CADD platform, please contact our team at info@vivabiotech.com.