- About Us
-
CRO Services
- PROTAC/Molecular Glue Services
- Protein Preparation and Ternary Complex Structure Determination
- PROTAC/Molecular Glues Screening
- PROTAC Ternary Complex Kinetics (SPR)
- PROTAC Degradation Assays and Ternary Complex Assays
- PROTAC Molecule Design and Synthesis
- ADME & PK/PD Studies of PROTAC Molecules
- AIDD/CADD PROTAC Design
- CDMO Services
- EFS Business
- News
- Careers
- Investor Relations
- Contact Us


Home / News / Events / Events detail
Guided by the Alumni Association of the Shanghai Institute of Materia Medica, Chinese Academy of Sciences, and hosted by the Cas Club, the "Pharma Digital Intelligence Innovation Development Seminar" was recently held at the Shanghai Institute of Materia Medica. Dr. Yue Qian, Executive Director of the Computational Chemistry and Artificial Intelligence Platform at Viva Biotech, was invited as a guest speaker, where she delivered a keynote speech titled "Applications of Artificial Intelligence in Drug Discovery."

Dr. Qian began by tracing the origins of computational chemistry, a field of research that has seen significant growth since the 1960s and is now playing a key role in various aspects of drug discovery and development. Then, she discussed the application of molecular dynamics in revealing drug mechanisms and protein conformational changes. Through molecular dynamics, drug developers can visualize the conformational changes of the target protein before and after its binding to small molecules. Using FGFR2 selective inhibitors as an example, she explained how molecular dynamics is applied in the study of protein conformations, providing critical guidance for the subsequent drug design.
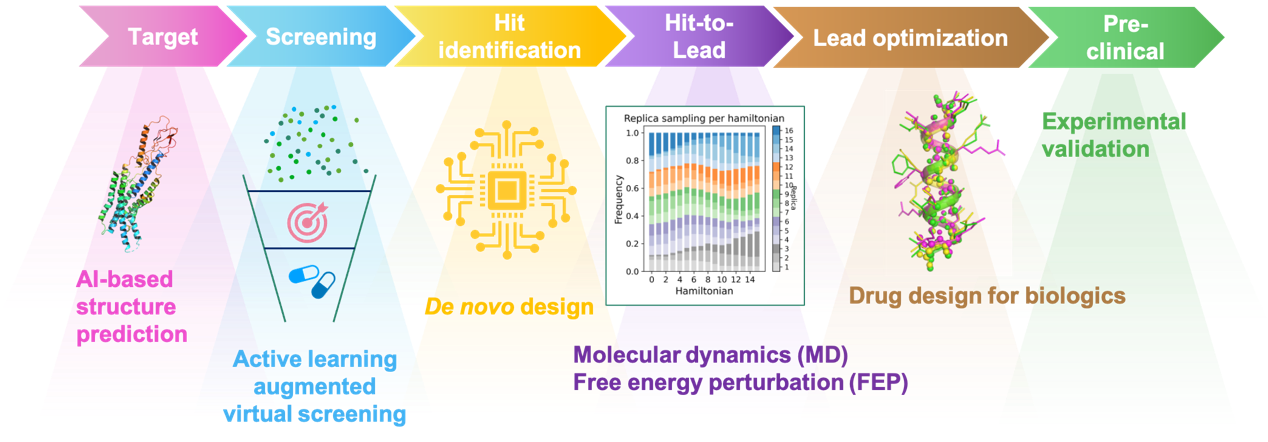
The computational-aided drug design workflow
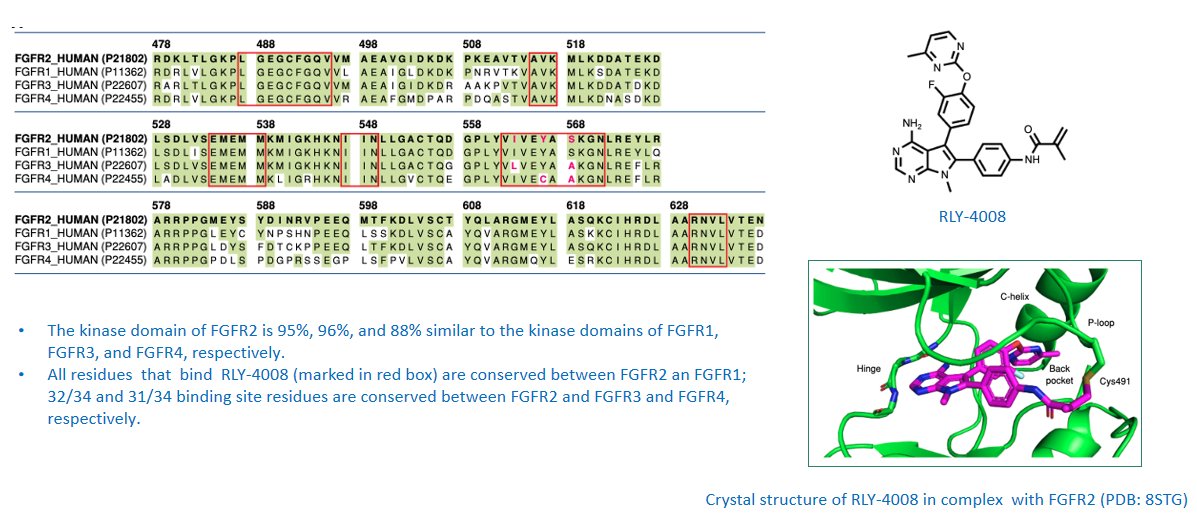
Case Study:design of FGFR2-selective inhibitor
Dr. Qian then discussed active learning augmented virtual screening, the first key element of which is obtaining protein structure and binding site information. When the protein structure is known but the binding site is unknown, molecular dynamics can help identify the binding modes or cryptic pockets by simulating the binding behavior of multiple fragment molecules along with the protein. Such practice lays the foundation of the subsequence virtual screening. The second element involves compound libraries. Traditional compound libraries are limited by chemical space. With the introduction of molecular generator, we can break the limitations and expand the chemical space to a much larger extent. For ultra-large compound libraries, maintaining high throughput while ensuring screening efficiency is challenging. To address this, Viva Biotech employs an active learning-accelerated virtual screening strategy that screens a random selection of compounds followed by iterative training and predicting, so that to capitalize the power of machine-learning along with the conventional docking. This active learning process itself is iteratively adaptive and ensures balance between accuracy and efficiancy.
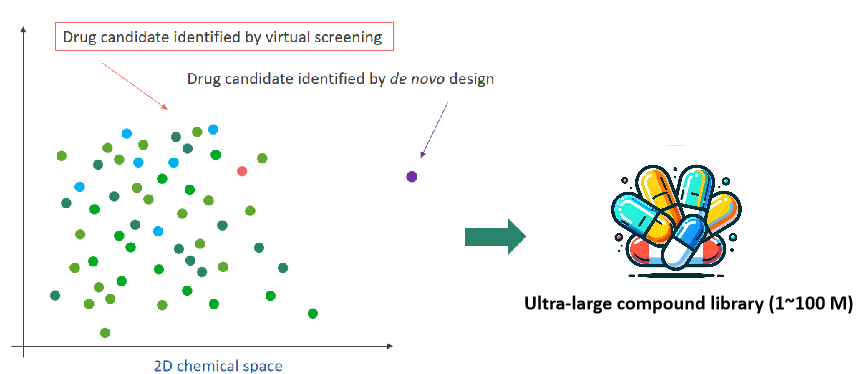
Molecular generation and active-learning based virtual screening
Next, Dr. Qian introduced the application of free energy perturbation (FEP) in lead optimization. There are mainly two approaches: relative free energy perturbation (RFEP) and absolute free energy perturbation (AFEP). RFEP considers the difference in free energy changes between two congeneric ligands in aqueous solution and protein complexes, which corresponds to their binding affinity differences. This method is suitable to scenarios where the molecular structures are similar to each other. AFEP, on the other hand, involves transforming the ligand of interest into a ghost state by turning off its interaction with the environment. In this way, we are able to overcome the significant structural change difficulties and avoid error cancellation. Based on the rigorous theory, free energy calculations can guide compound design and accelerate the lead optimization process. Dr. Qian emphasized that Viva Biotech's in-house FEP platform achieves the level of accuracy matching best-in-class software across a number of benchmark systems. In a recent paper, "Adaptive Lambda Schemes for Efficient Relative Binding Free Energy Calculation," Dr. Qian and coworkers further improved the efficiency and stability of relative binding free energy calculations.
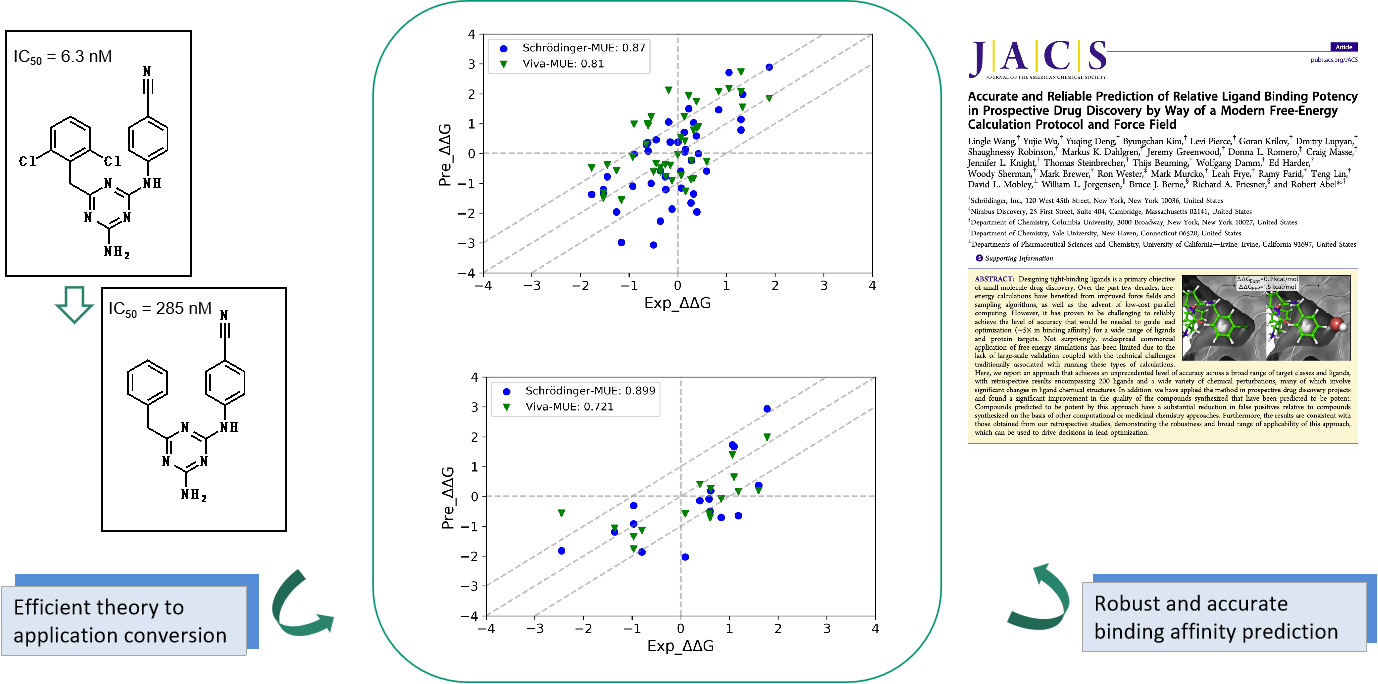
Comparison of FEP platform between Viva Biotech and Schrödinger
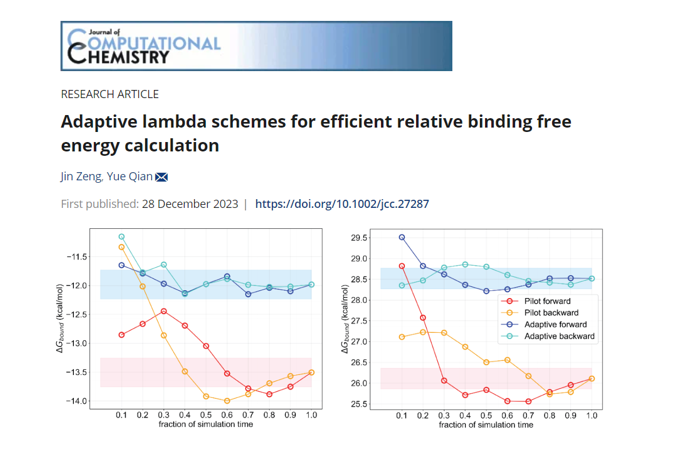
Source: J. Comput. Chem.
In addition to small molecule applications, Dr. Qian shared experiences with computational-aided drug design (CADD) in biologics drug development, particularly in antibody affinity maturation such as developing ADCs and XDCs. While de novo design is a popular topic in academia, antibody-related work is still in its early stage. Accurate antibody design requires both physics-based models and AI-based models. Dr. Qian detailed an affinity maturation case where researchers first modeled the antibody to obtain the antigen-antibody complex structure, using a combination of physical models and deep learning mainly for CDR optimization. The team then evaluated protein-protein interactions, performed energy analysis, and identified key residues for mutation. Besides affinity, antibody developability indices were also assessed. In this case, 20 sequences were initially screened experimentally, with 7 showing comparable activity to the wild type and 1 showing a 4-fold improvement. A second round of design and testing based on experimental results led to a sequence with a 7-fold improvement in activity. This combined computational and experimental approach significantly enhanced the success rate and speed of antibody screening, complementing traditional phage display screening. She also shared a systematic evaluation of the FDA-approved antibody developability profile, focusing on CDR and Fv regions, considering length, hydrophobicity, viscosity, solubility, stability, and surface charge, to determine developability indices.
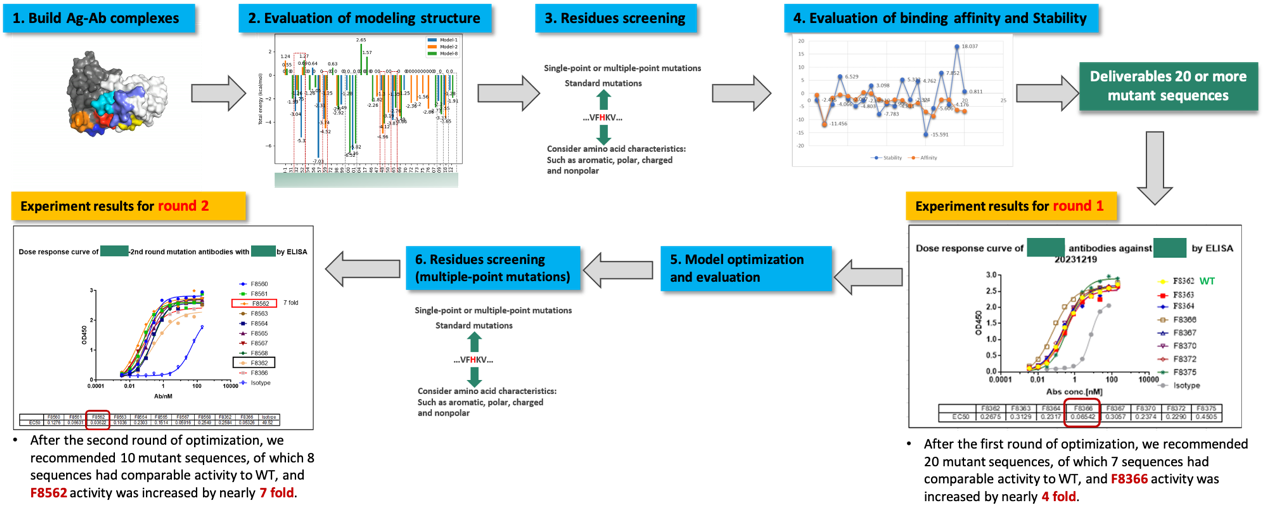
Case Study: Antibody affinity maturation
Finally, Dr. Qian emphasized that techniques such as deep learning, molecular dynamics, and free energy perturbation require substantial computational power. She highlighted that Viva Biotech currently utilizes a local supercomputing center that provides sufficient computational resources. Moreover, the company remains committed to optimizing algorithms and accumulating high-quality data to further advance drug design efforts.
Copyright © Viva Biotech All Rights Reserved. 沪ICP备19036061号
- About Us
-
CRO Services
BackCRO ServicesService & Technology
- CDMO Services
- EFS Business
- News
- Careers
- Investor Relations
- Contact Us